# Three-line Summary #
- We present a network and training strategy that relies on the strong use of data augmentation to use the available annotated samples more efficiently.
- The architecture consists of a contracting path to capture context and a symmetric expanding path that enables precise localization.
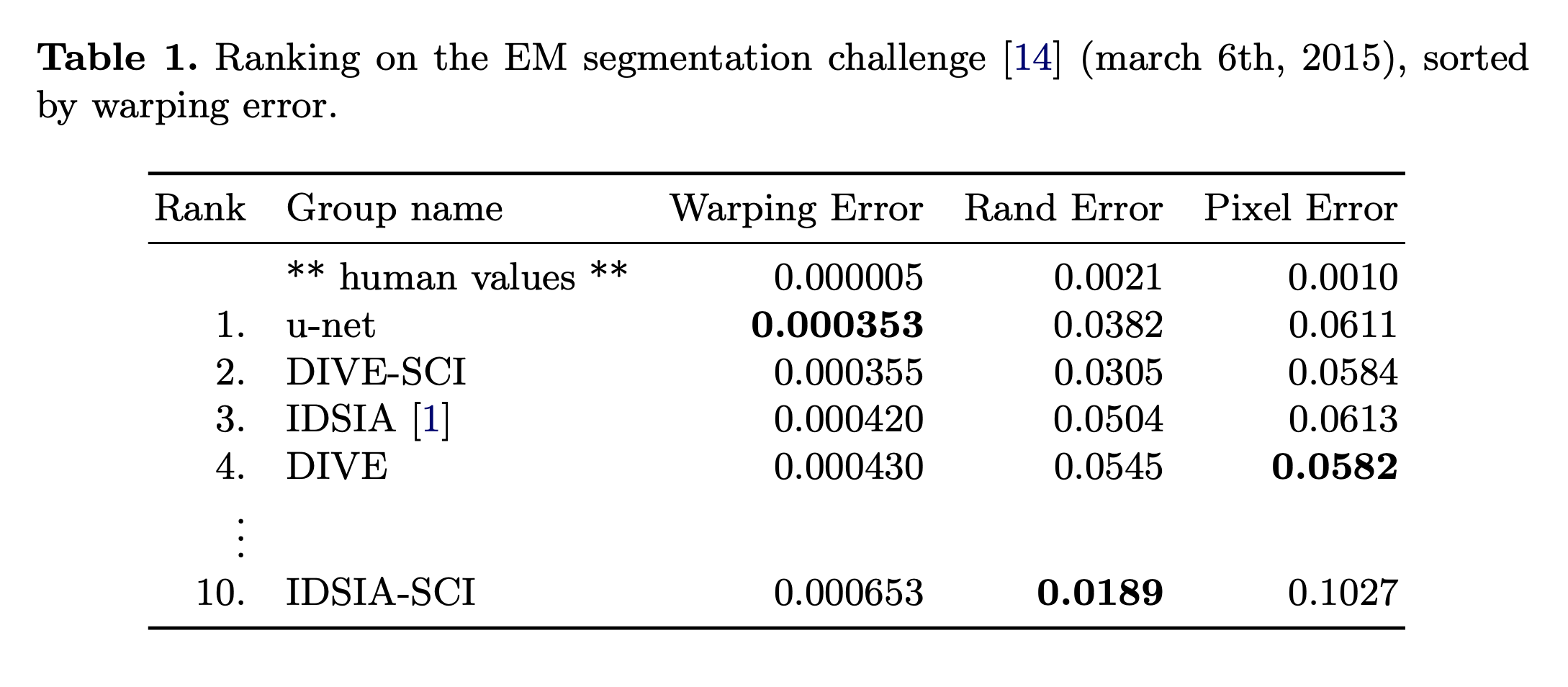
- Using the network trained on transmitted light microscopy images, we won the ISBI cell tracking challenge 2015 in these categories by a large margin.
# Detail Review #
1. Introduction
- In many visual tasks, especially in biomedical image processing, the desired output should include localization, i.e., a class label is supposed to be assigned to each pixel.
- [Ciresan et al., 2012] trained a network in a sliding-window setup to predict the class label of each pixel by providing a local region (patch) around that pixel as input
- [Ciresan et al., 2012] drawback 1. it is quite slow because the network must be run separately for each patch.
- [Ciresan et al., 2012] drawback 2. there is a trade-off between localization accuracy and the use of context (Larger patches -> reduced localization accuracy, small patches -> see only little context).
- We modify and extend FCN (Long et al., 2014) architecture; the main idea in FCN is to supplement a usual contracting network with successive layers, where pooling operators are replaced by upsampling operators.
- One important modification in our architecture is that in the upsampling part, we also have a large number of feature channels, which allow the network to propagate context information to higher resolution layers.
- As a consequence, the expansive path is more or less symmetric to the contracting path and yields a u-shaped architecture.
- This strategy allows the seamless segmentation of arbitrarily large images by an overlap-tile strategy.
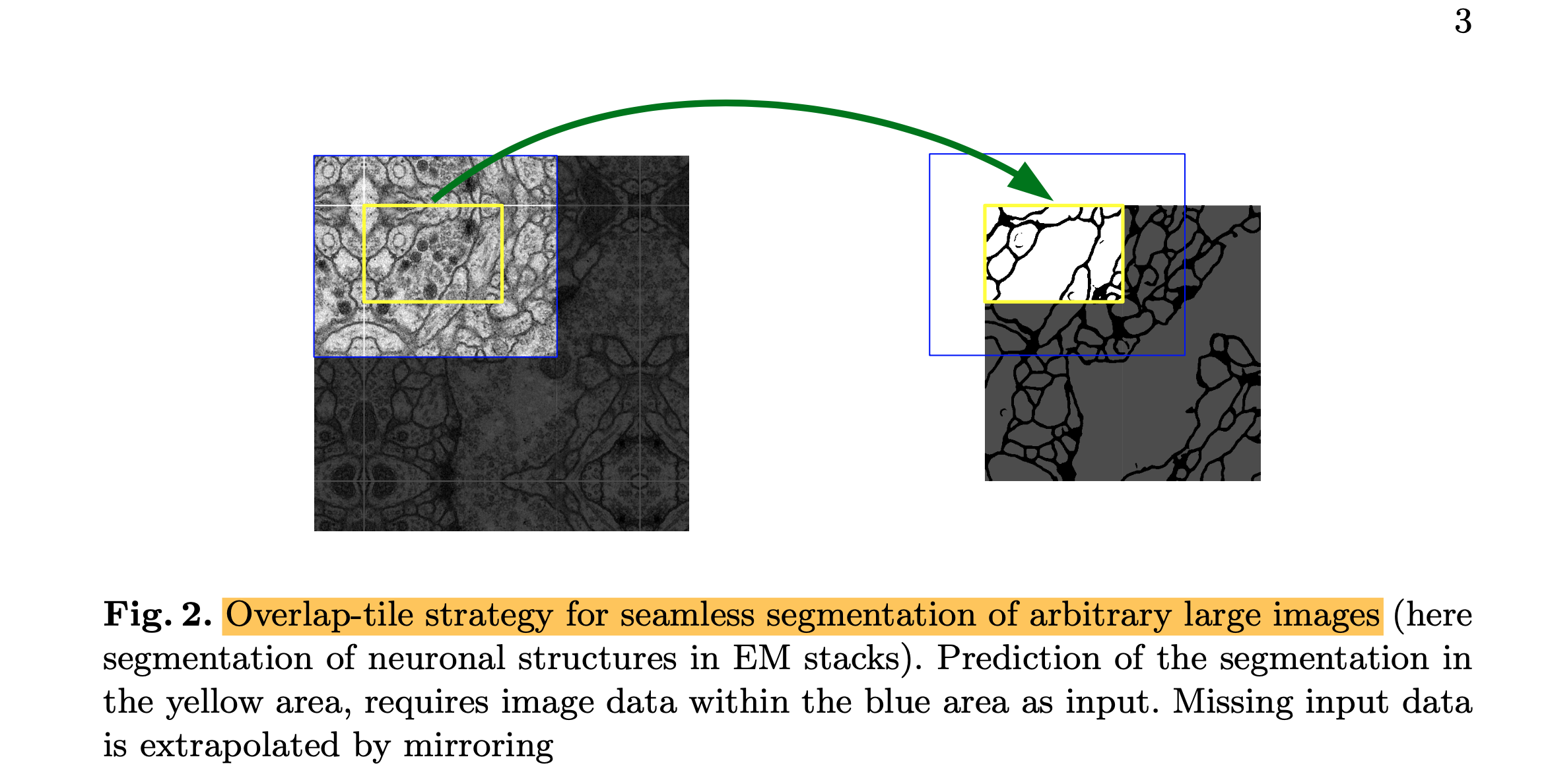
- As for our tasks, there are very little training data available, we use excessive data augmentation by applying elastic deformations to the available training images.
- This is particularly important in biomedical segmentation since deformation used to be the most common variation in tissue, and realistic deformations can be simulated efficiently.
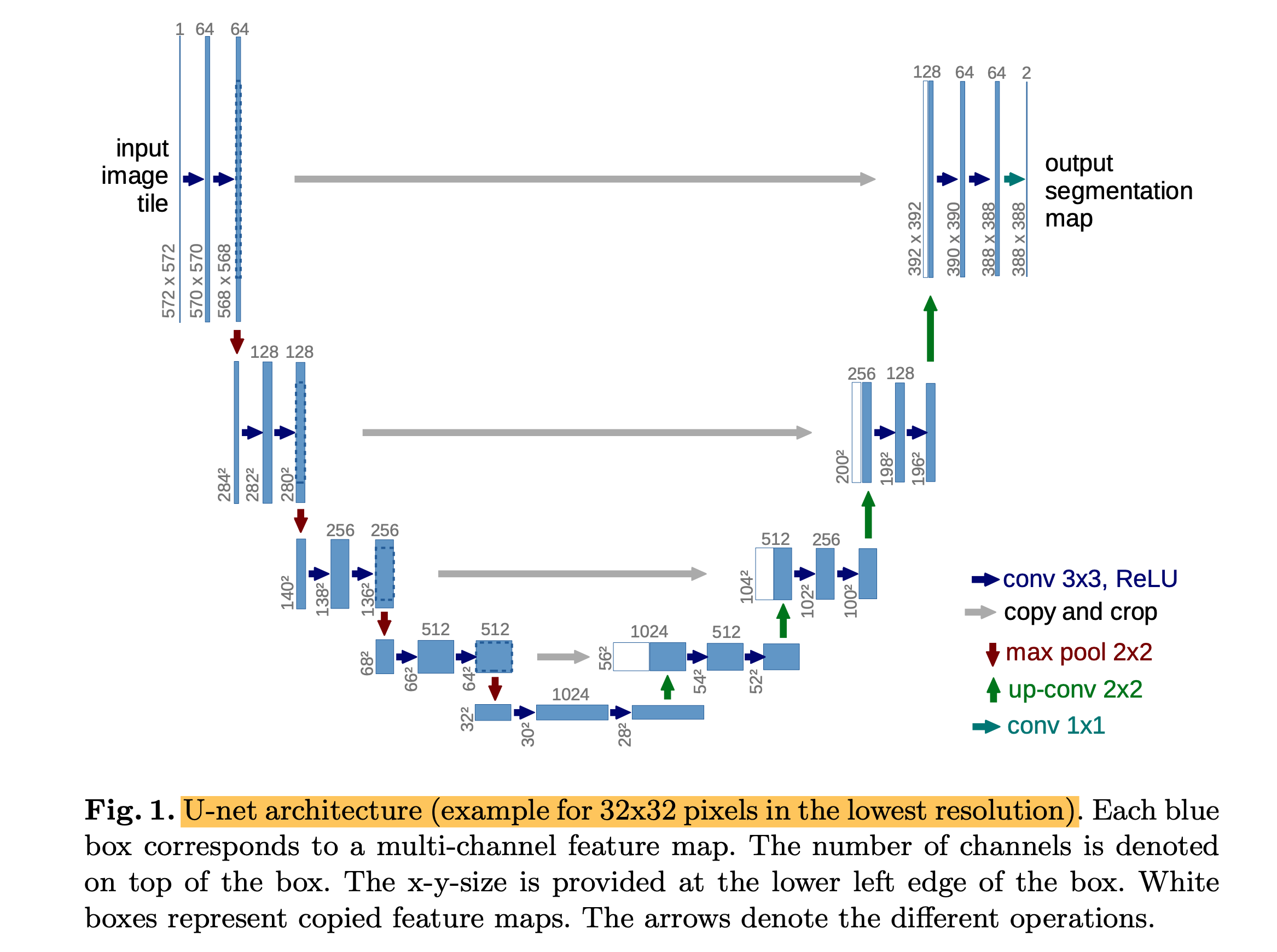
2. Architecture
- U-Net architecture consists of a contracting path (left side) and an expansive path (right side).
- The contracting path follows the typical architecture of a convolutional network.
- Every step in the expansive path consists of an upsampling of the feature map followed by a 2x2 convolution (up-convolution), a concatenation with the correspondingly cropped feature map from the contracting path.
- At the final layer, a 1x1 convolution is used to map each 64-component feature vector to the desired number of classes.
- The input images and their corresponding masks are used to train the network with the stochastic gradient descent, and the energy function is computed by a pixel-wise softmax over the final feature map combined with the cross entropy loss function.
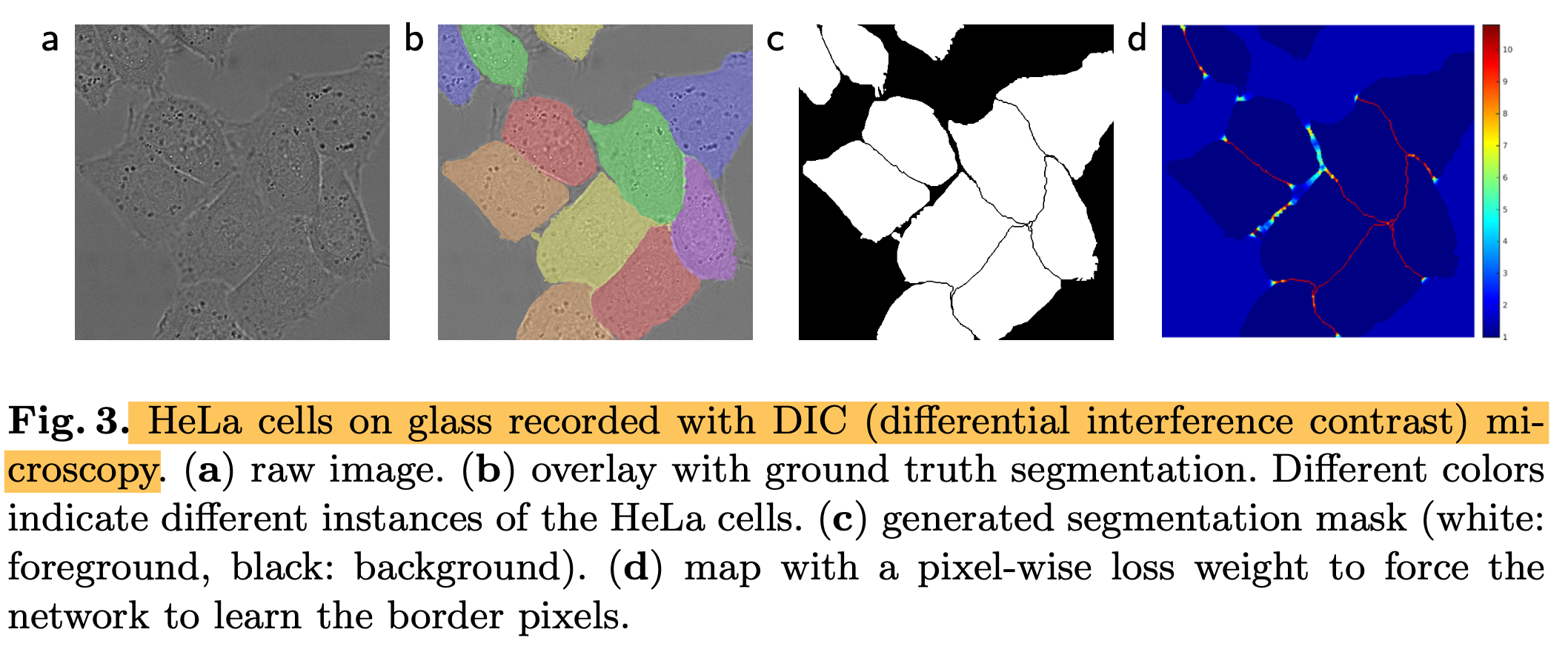
- we pre-compute the weight map for each ground truth segmentation to compensate the different frequency of pixels from a certain class in the training dataset and to force the network to learn the small separation borders that we introduce between touching cells.
- Data augmentation is essential to teach the network the desired invariance and robustness properties when only a few training samples are available.
- we generate smooth deformations using random displacement vectors on a coarse 3 by 3 grid.
3. Experiments
- We demonstrate the application of the u-net to three different segmentation tasks.
- (1) The segmentation of neuronal structures in electron microscopic recordings (fig 2).
- The dataset is provided by the EM segmentation challenge that was started at ISBI 2012.
- The evaluation is done by thresholding the map at 10 different levels and computation of the "wrapping error", the "Rand error" and the "pixel error".
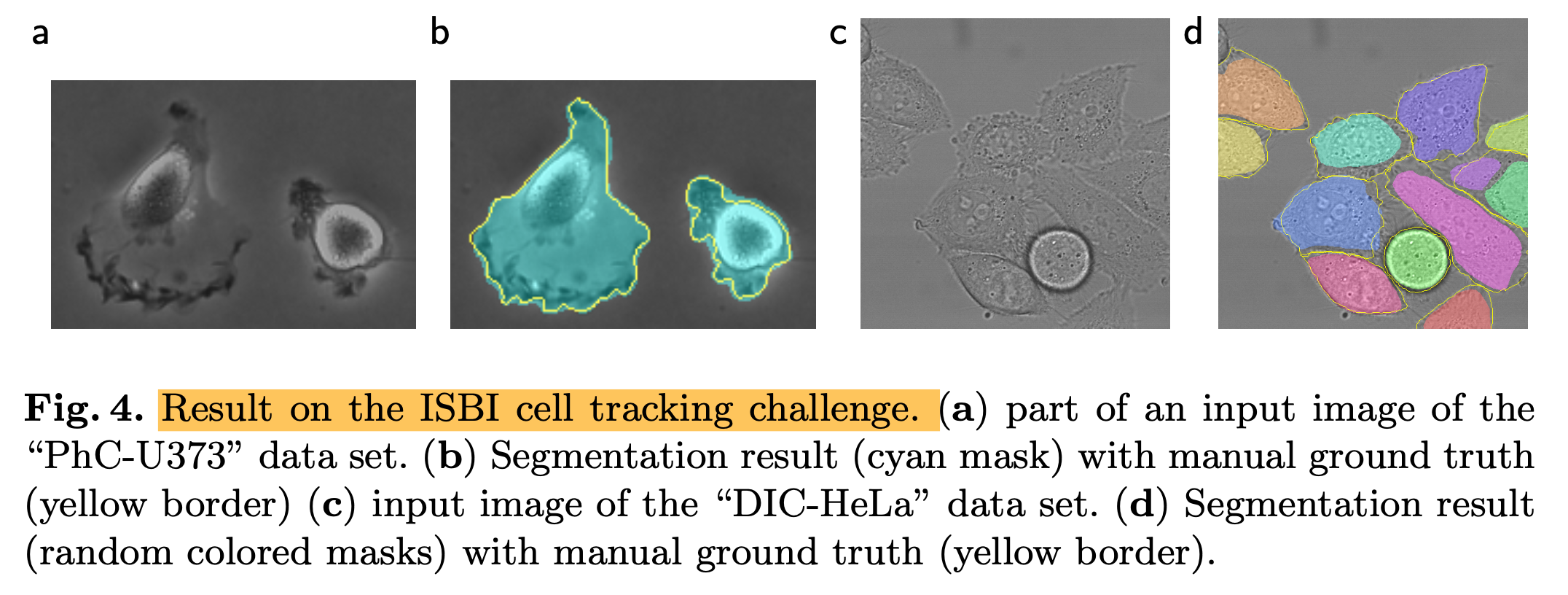
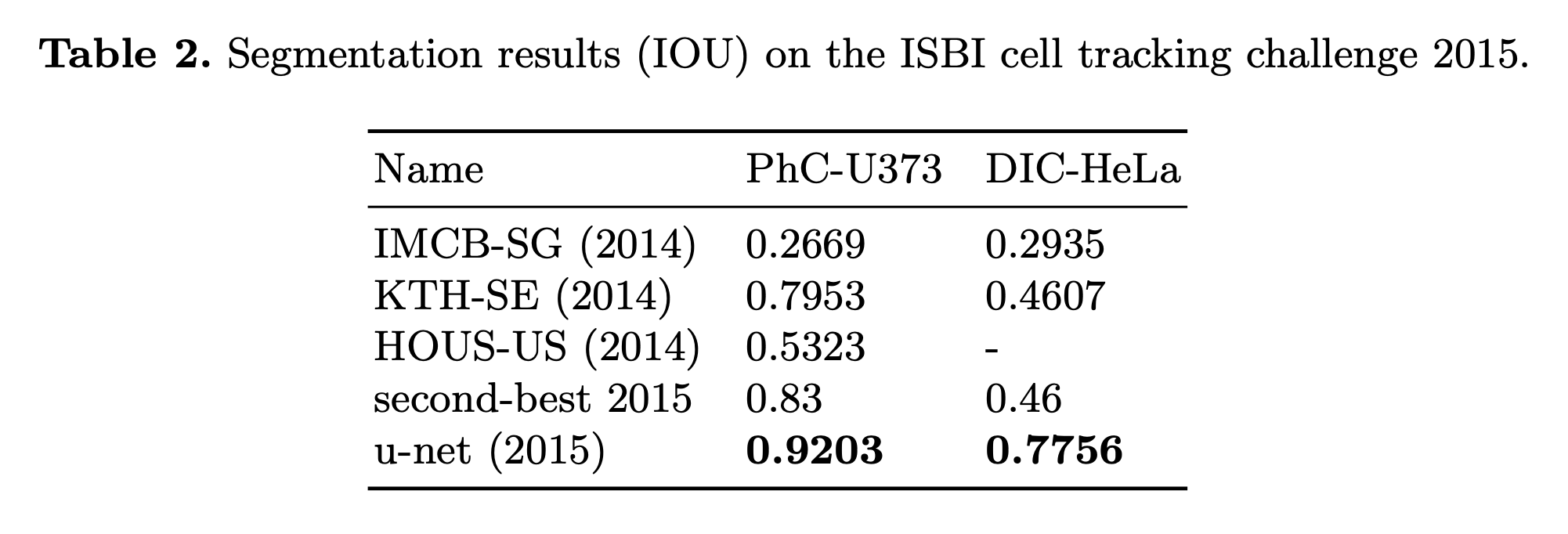
- We also applied the u-net to a cell segmentation task in light microscopic images that segmentation task is part of the ISBI cell tracking challenge 2014 and 2015
- (1) PhC-U373 -> IOU = 92%
- (2) DIC-HeLa -> IOU = 77.5%
* Reference: Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
- Ciresan et al., 2012: https://proceedings.neurips.cc/paper/2012/hash/459a4ddcb586f24efd9395aa7662bc7c-Abstract.html
Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images
Requests for name changes in the electronic proceedings will be accepted with no questions asked. However name changes may cause bibliographic tracking issues. Authors are asked to consider this carefully and discuss it with their co-authors prior to reque
proceedings.neurips.cc
728x90
728x90




댓글