# 세줄 요약 #
- With the rapid development of image scanning techniques and visualization software, whole slide imaging (WSI) is becoming a routine diagnostic method.
- Deep learning-based pathology image segmentation has become an important tool in WSI analysis because that algorithms such as fully convolutional networks stand out for their accuracy, computational efficiency, and generalizability.
- In this review, we are to provide quick guidance for implementing deep learning into pathology image analysis and to provide some potential ways of further improving segmentation performance.
# 상세 리뷰 #
1. Introduction
- Convolutional neural network (CNN) introduced, which have been widely used for pathology image analysis, such as WSI patch classification, tumor region and metastasis detection.
- To perform image segmentation for large data (eg, whole slide pathology image),
- the image is divided into many small patches and classify those patches by CNN,
- then all patches in the same class are combined into one segment area.
- But image segmentation from classifying patch method demands a substantial computing time and memory (to overcome segmentation resolution, generate too many patches),
- So this paper provides the segmentation deep-learning algorithms that refer to semantic or instance segmentation algorithms
- that are more computationally efficient in pixel classification & extract detailed image information.
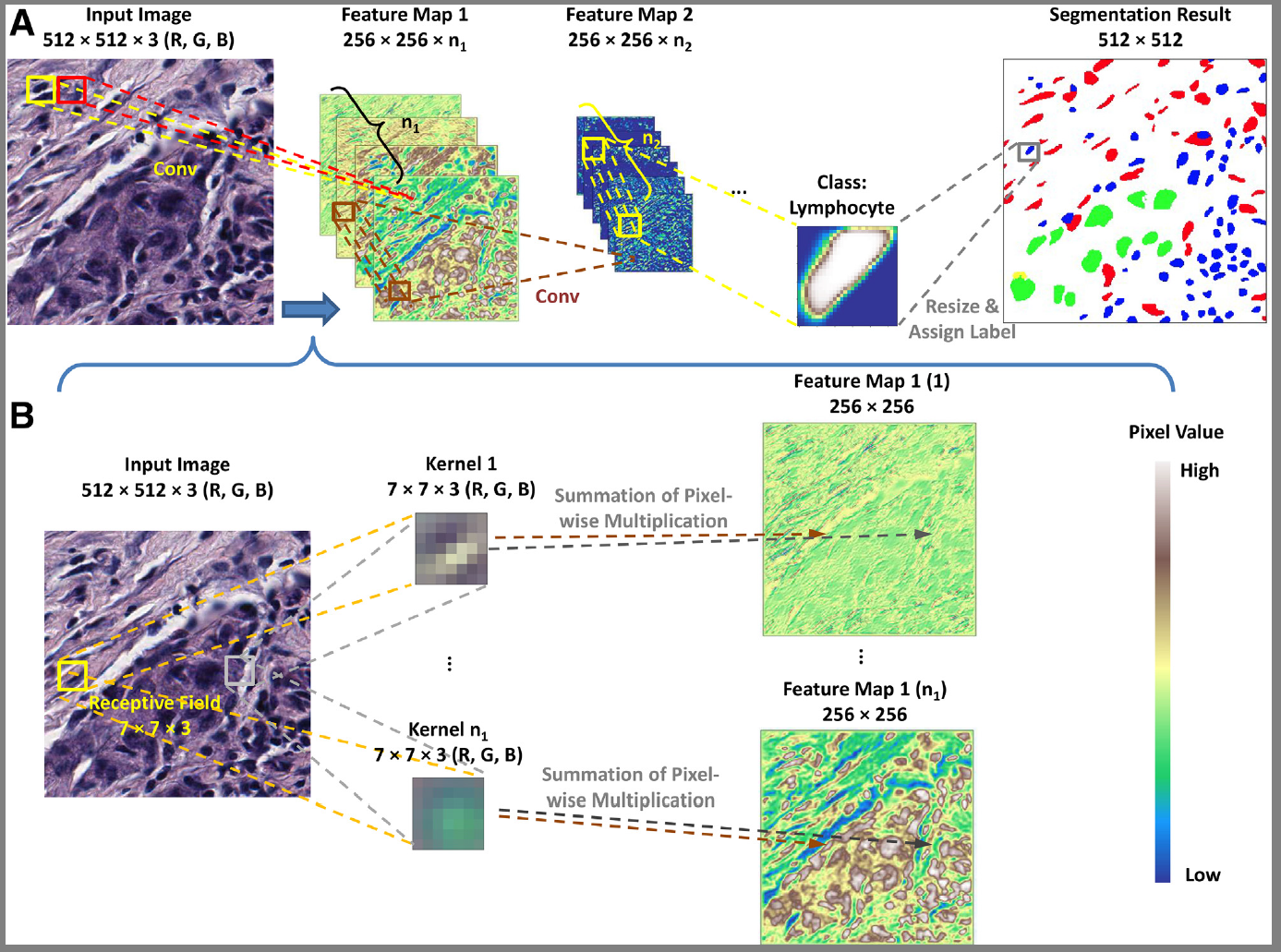
2. Dataset and Preprocessing
- Pathology images are usually as large as giga-pixels, so the pathology images should first be chopped into small patches that are resized or padded at the same size before being fed to the neural network.
- Image normalization ensure that different features have a similar effect on the response, because normalization helps accelerate convergence in a step-wise gradient algorithm
- rescale: [0, 1] or [-1, 1]
- standardization: mean = 0, varience = 1
- Because pathology images may look very different due to different staining conditions and slide thickness,
- it is important to use color augmentation to mimic practical differences and ignore systematic biases by adding a random mean and multiplying a random variation to each channel of each image.
3. Model Selection and Construction
- Model Selection:
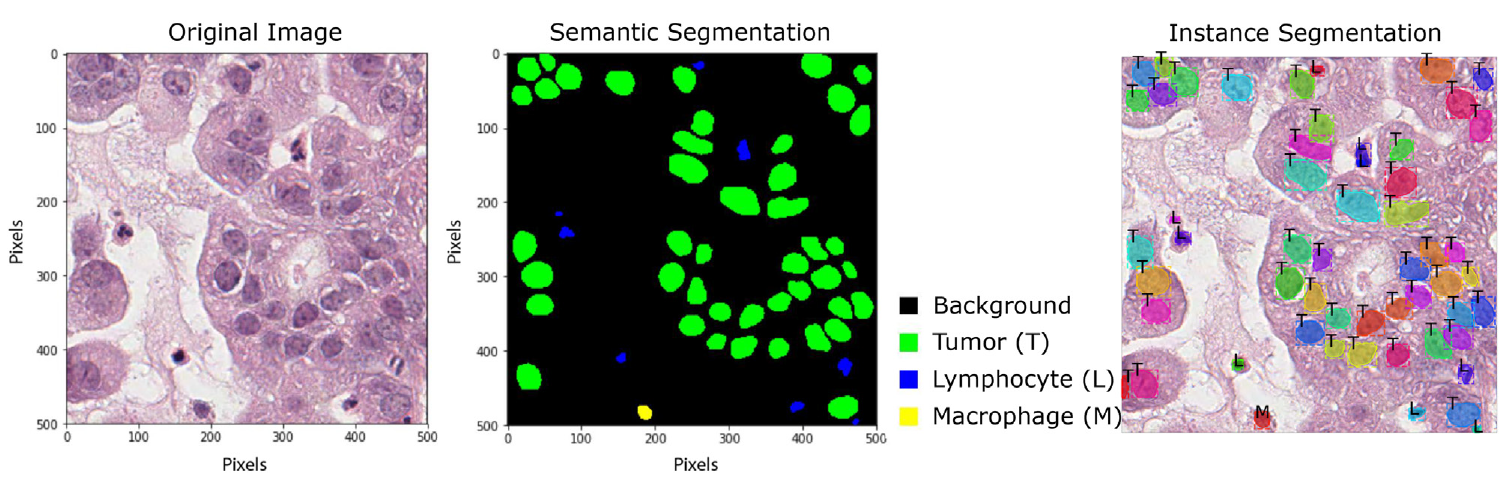
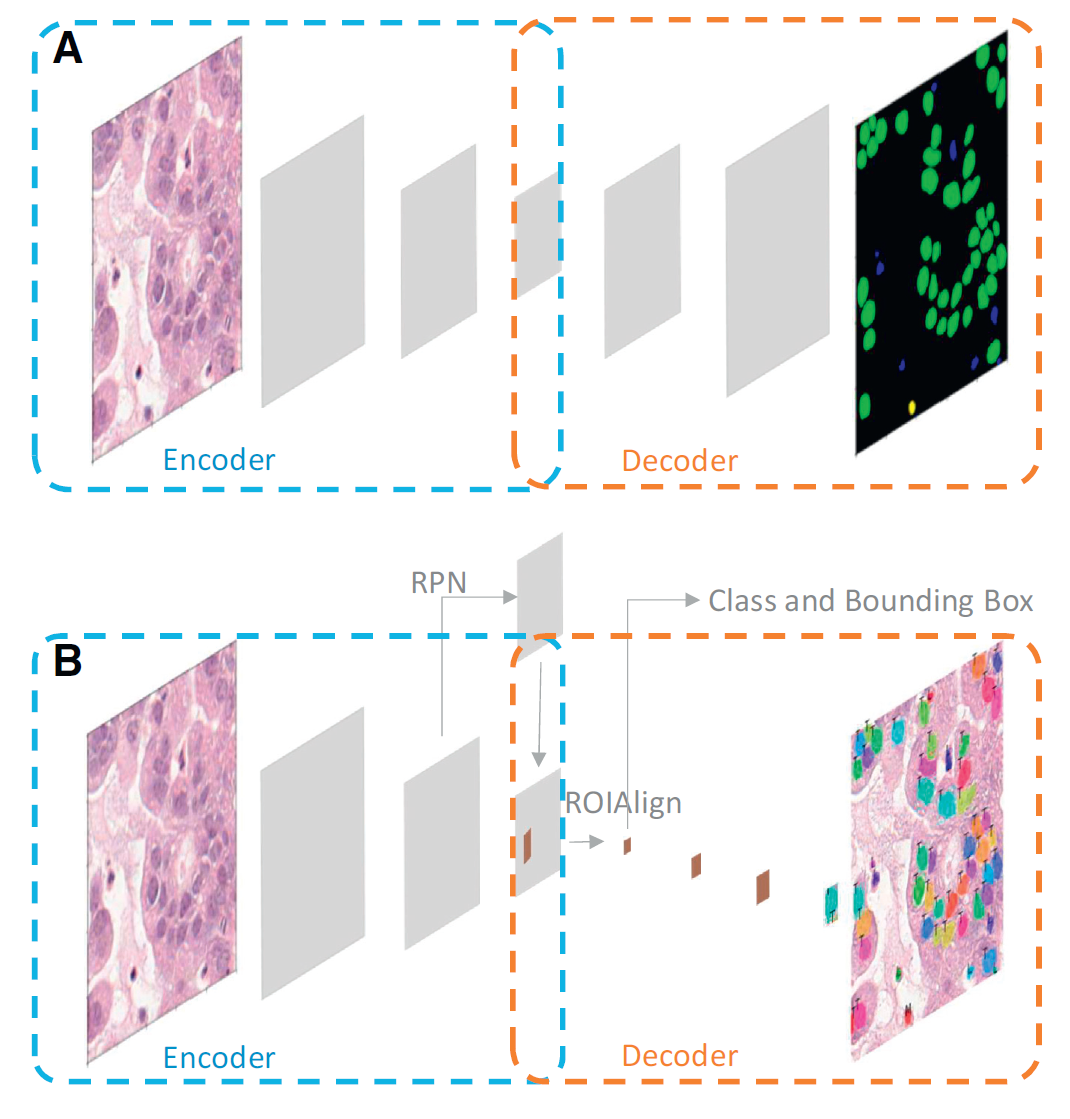
- 1) semantic segmentation is to segment image parts with a different meaning (ex. FCN, U-Net, DeepLab),
- 2) instance segmentation, which detects the region of interest (ROI) for each instance first, then classification and segmentation are applied to the same ROI in parallel (ex. Mask R-CNN).
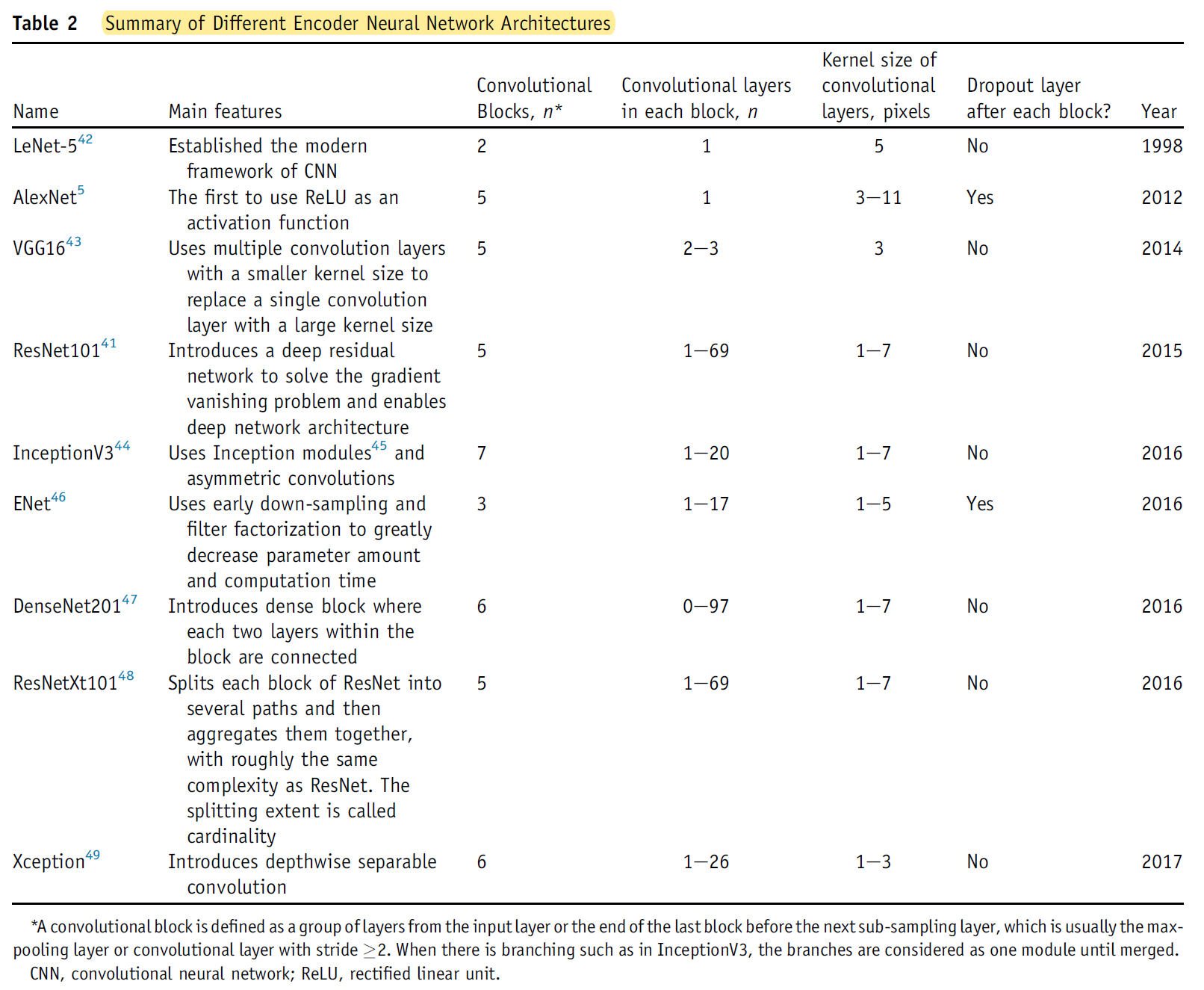
- Finally, choosing a proper backbone network structure is critical for successful approximation (see Table 2).
- Loss Function:
- 1) for semantic segmentation, the most common loss function is pixel-wise cross-entropy between the network outputs and the true segmentation annotations,
- 2) For instance segmentation, the losses are composed of three parts
- categorical classification cross-entropy
- bound-box regression L1 loss
- pixel-wise binary cross-entropy).
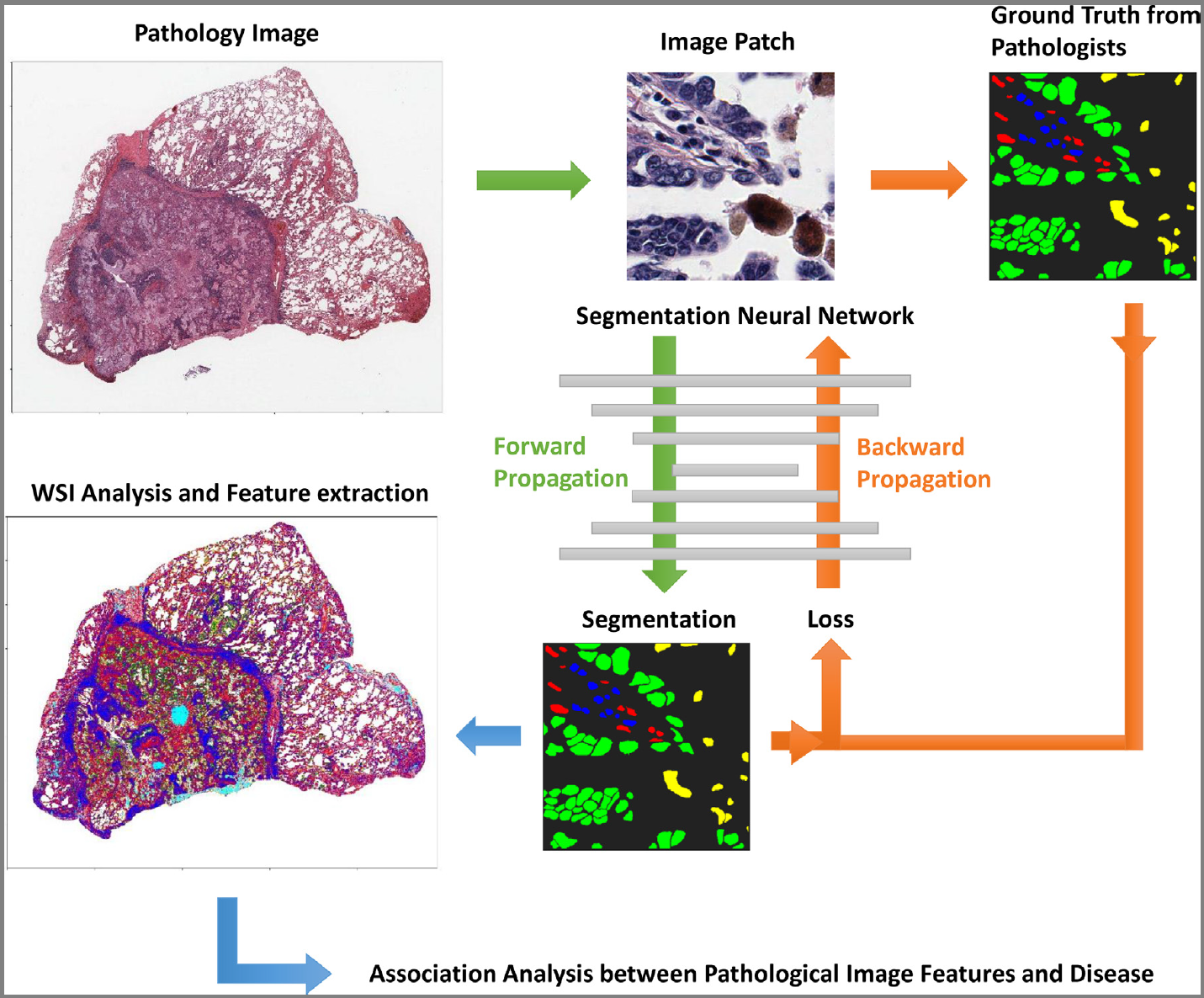
- Training phase:
- The training phase is a process to update model parameters and is composed of alternating forward and backward propagations,
- but the loss function of neural networks is usually not convex,
- so we choose
- transfer learning (used backbone and using pre-trained weights as initial parameters)
- Autoencoder (reconstruct the inputs before supervised learning).
* Reference: Wang, Shidan, et al. "Pathology image analysis using segmentation deep learning algorithms." The American journal of pathology 189.9 (2019): 1686-1698.




댓글