# 세줄 요약 #
- The development of decision support systems for pathology and their deployment in clinical practice have been hindered by the need for large manually annotated datasets.
- We present a multiple instance learning-based deep learning system that uses only reported diagnoses as labels for training.
- Tests on prostate cancer, basal cell carcinoma and breast cancer metastases to axillary lymph nodes resulted in areas under the curve above 0.98 for all cancer types.
# 상세 리뷰 #
1. Introduction
1.1. Digital Pathology
- In recent years has digital pathology emerged as a potential new standard of care where glass slides are digitized into whole slide images (WSIs) using digital slide scanners.
- But computational pathology has to face additional challenges
- (1) The lack of large annotated datasets.
- (2) pathology images are tremendously large (470 WSIs contain roughly the same number of pixels as the entire ImageNet dataset)
- At that problem, reliance on expensive and time-consuming, manual annotations is impossible.
- Proposing a new framework for training classification models at a very large scale without the need for pixel-level annotations.
1.2. Dataset
- We collected three datasets in the field of computational pathology
- (1) A prostate core biopsy dataset: 24,859 slides
- (2) A skin dataset: 9,962 slides
- (3) A breast metastasis to lymph nodes dataset: 9,984 slides
- We propose to use the slide-level diagnosis, to train a classification model in a weakly supervised manner.
- To be more specific, the slide-level diagnosis casts a weak label on all tiles within a particular WSI.
- if the slide is negative: all of its tiles must also be negative and not contain tumor.
- if the slide is positive: it must be true that at least one of all of the possible tiles contains tumor.
- To be more specific, the slide-level diagnosis casts a weak label on all tiles within a particular WSI.
1.3. Method
- Multiple Instance Learning (MIL)
- widely applied in many machine learning domains, including computer vision.
- weakly supervised WSI classification rely on deep learning models trained under variants of the MIL assumption.
- A two-step approach,
- (1) A classifier is trained with MIL at the tile level
- (2) The predicted scores for each tile within a WSI are aggregated,
- by combining (pooling) their results with various strategies.
- by learning a fusion model.
- MIL to train deep neural networks
- Used in a recurrent neural network (RNN) to integrate the information across the whole slide and report the final classification result.
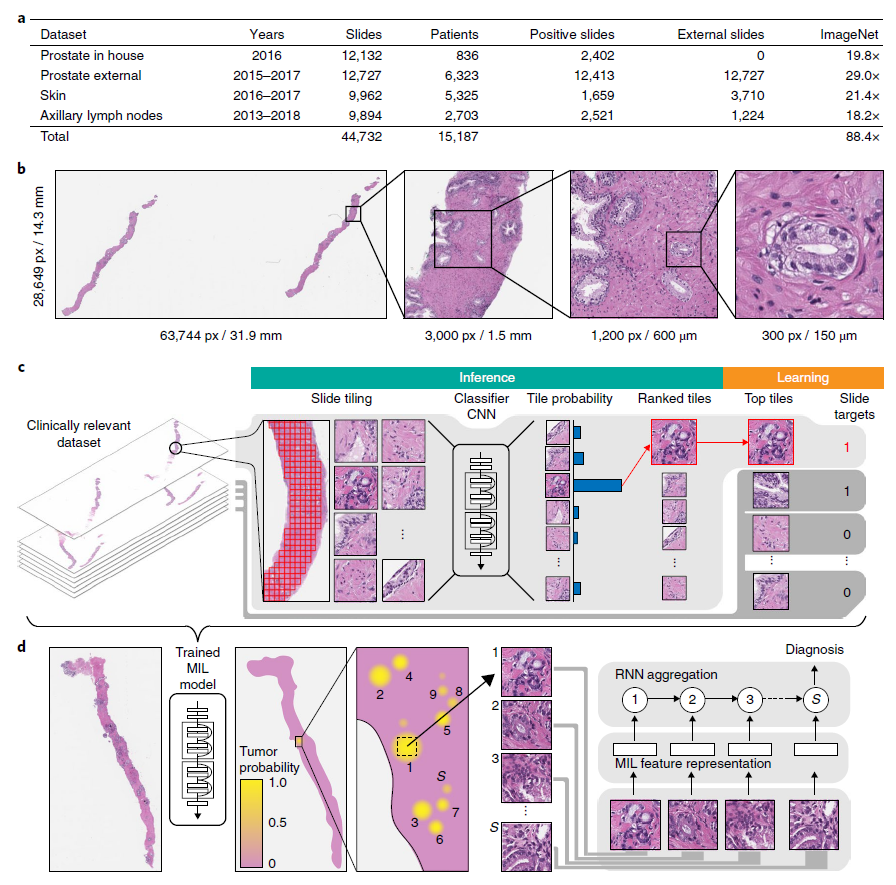
- (a) Description of the datasets.
- This study is based on a total of 44,732 slides from 15,187 patients across three different tissue types: prostate, skin, axillary lymph nodes.
- (b) Hematoxylin and Eosin (H&E) slide of biopsy showing prostatic adenocarcinoma.
- The diagnosis can be based on very small foci of cancer that account for < 1% of the tissue surface.
- (c) The MIL training procedure includes a full inference pass through the dataset, to rank the tiles according to their probability of being positive, and learning on the top-ranking tiles per slide.
- (d) Slide-level aggregation with a recurrent neural network (RNN).
- The S most suspicious tiles in each slide are sequentially passed to the RNN to predict the final slide-level classification.
2. Result
2.1. Test performance of DNN models (ResNet34) trained with MIL

- (a) Best results were achieved on the prostate dataset (n = 1,784),
- AUC = 0.989 at 20x magnification
- (b) For BCC (Basal Cell Carcinoma) (n = 1,575),
- AUC = 0.990 at 5x magnification
- (c) The breast metastasis detection task (n = 1,473),
- AUC = 0.965 at 20x magnification
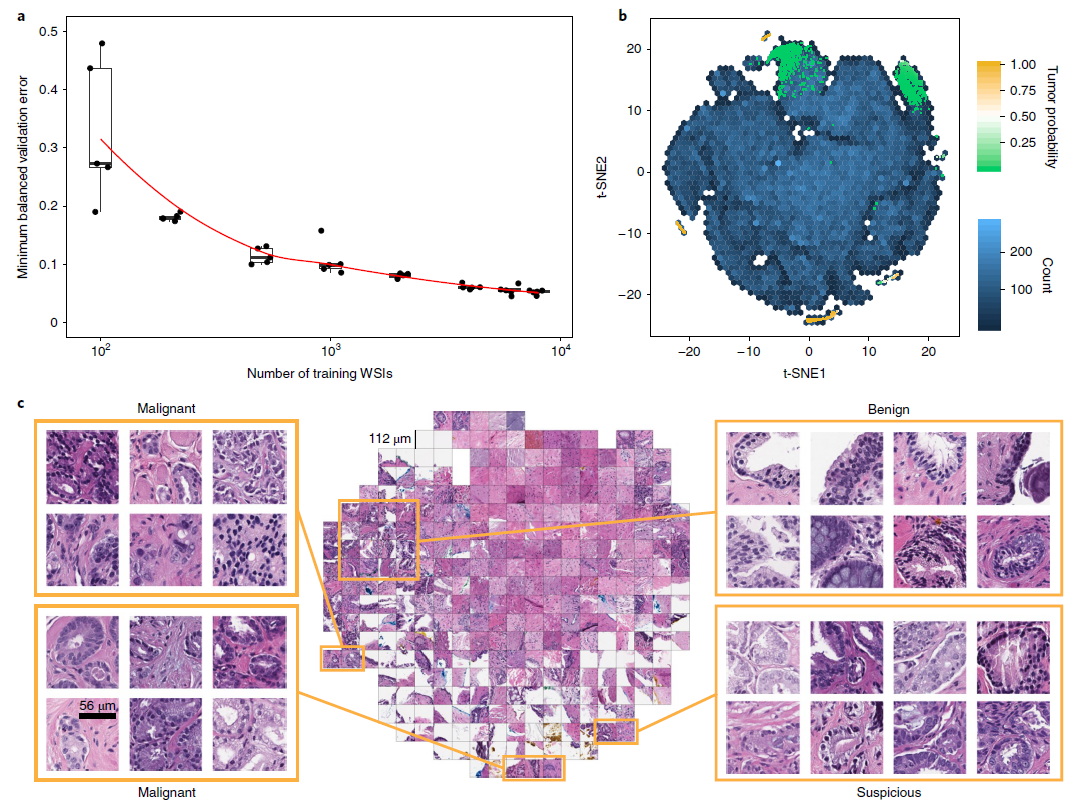
- (a) Dataset size plays an important role in achieving clinical-grade MIL classification performance.
- Training of ResNet34 was performed with datasets of increasing size
- Validation set = 2,000 slides, Training sets = 100, 200, 500, 1000, 2000, 4000, 6000, 8000 slides
- A large number of slides are necessary for generalization of learning under MIL assumption.
- Training of ResNet34 was performed with datasets of increasing size
- (b) A ResNet34 model trained at 20x was used to obtain the feature embedding before the final classification layer for a random set of tiles in the test set (n = 182,912).
- The embedding was reduced to two dimensions with t-SNE and plotted using a hexagonal heat map.
- (c) Tiles corresponding to points in the two-dimensional t-SNE space were randomly sampled from different regions.
- Abnormal glands: clustered together on the bottom and left sides of the plot.
- Suspicious glands (tumor probability ~ 0.5): clustered on the bottom region of the plot.
- Normal glands: clustered on the top left region of the plot.
2.2. Weakly supervised learning Result Analysis

- The performances of the models trained at 20x magnification on the respective test datasets were measured in terms of AUC for each tumor type.
- (a) For prostate cancer (n = 1,784): AUC = 0.991
- The MIL-RNN model significantly outperformed the model trained with MIL alone.
- (b) For BCC model (n = 1,575): AUC 0.988
- (c) For breast metastases detection (n = 1,473): AUC = 0.966
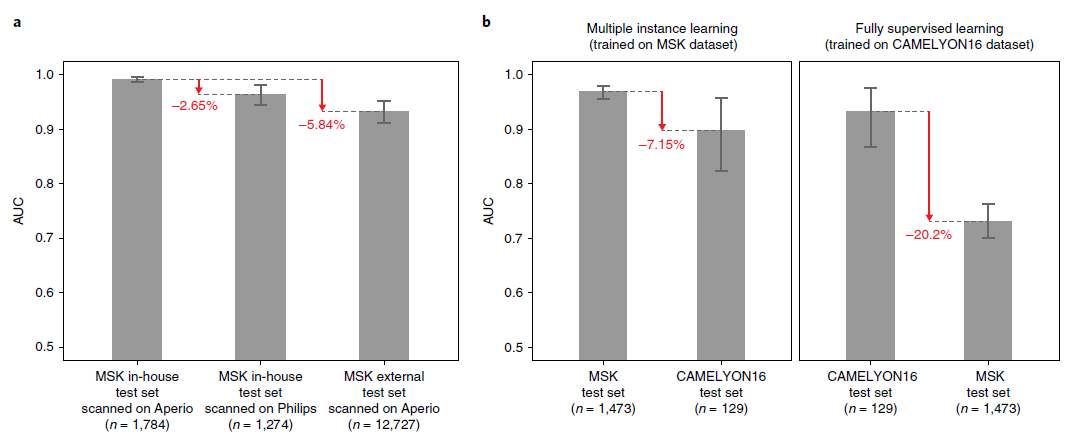
- The generalization performance of the proposed prostate and breast models were evaluated on different external test sets.
- (a) Results of the prostate model trained with MIL on MSK (Memorial Sloan Kettering Cancer Center) in-house slides and tested on:
- 1) The in-house test set (n = 1,784) scanned on Aperio
- 2) The in-house test set (n = 1,274) scanned on Philips
- 3) external slides submitted to MSK for consultation (n = 12,727)
- (b) Comparison of the proposed MIL approach with state-of-the-art fully supervised learning for breast metastasis detection in lymph nodes
- Left, the model was trained on MSK data with our proposed method (MIL-RNN)
- The MSK breast data test set (n = 1,473): AUC = 0.965
- The test set of the CAMELYON16 challenge (n = 129): AUC = 0.899 (* decrease in AUC of 7%)
- Right, the model was trained CAMELYON16 data with a fully supervised model.
- The CAMELYON16 test set (n = 129): AUC = 0.930
- The MSK test set (n = 1,473): AUC 0.727 (* its performance drops by over 20%)
- Left, the model was trained on MSK data with our proposed method (MIL-RNN)
2.3. Conclusion
- These results illustrate that current deep learning models,
- Trained on small datasets, pixel-wise labels,
- Not able to generalize to clinical-grade, real-world data.
- These results also show that weakly supervised approaches,
- A clear advantage over conventional fully supervised learning
- They enable training on massive, diverse datasets without the necessity for data curation
# Reference: Campanella, Gabriele, et al. "Clinical-grade computational pathology using weakly supervised deep learning on whole slide images." Nature medicine 25.8 (2019): 1301-1309.




댓글