# Three-line Summary #
- Artificial Intelligence based on Deep Learning (DL) is opening new horizons in biomedical research and promises to revolutionize the microscopy field.
- We introduce recent developments in DL applied to microscopy in a manner accessible to non-experts.
- We discuss how DL shows an outstanding potential to push the limits of microscopy, enhancing resolution, signal, and information content in acquired data.
# Detail Review #
1. Introduction
- In the early 2010s, one type of machine learning, Deep Learning (DL), based on so-called neural networks (NNs), became increasingly prominent as a tool for image classification with super-human capabilities.
- NN is initially presented with a large set of paired input and desired output called the training dataset, from which it learns how to map each input into its corresponding desired output.
- Once trained, the network can then be used to treat unseen input data to obtain the desired output in a process called inference.
- The success of NNs in image recognition is thus closely linked to the exponential increase in the computational power of processing units, notably Graphical Processing Units (GPUs), and the rapid growth in the availability of large datasets since the 2000s.
- Although NNs were first envisioned in the 1950s, it took decades and the introduction of back-propagation until the first NN reached significant performances in pattern recognition tasks in the late 1980s.
- In 2012, the first GPU-enabled NN called AlexNet vastly outperformed the competition at the ImageNet image classification challenge, a seminal breakthrough for the AI field.
- In the latter field, NN applications include automated, accurate classification and segmentation of cell images, extraction of structures from label-free microscopy imaging (artificial labeling), and most recently image restoration (denoising and super-resolution).
- Here, we give non-specialist readers an overview of the potential of NNs in the context of some of the major challenges of microscopy.
- We also discuss some of their current limitations and give an outlook on possible future applications in microscopy.
- We recommend reviews that comprehensively discuss the application of AI in biomedical sciences and computational biology.

2. Neural network learning
- NNs are complex networks of connected 'neurons' arranged in 'layers', and each layer then provides a new representation of the data to the next layer with growing levels of abstraction (so the deeper network can extract more complex information in data).
- An important form of NN, especially for tasks involving feature recognition in image data are convolutional neural networks (CNNs).
- CNN contains convolutional layers (extract image features on the input image) and pooling layers (reduce the number of pixels in the image and therefore simplify the feature representations).
- This combination of successive feature extractions and data shrinkage leads to a simplified version of the input image, which the network learns to associate to the desired output.
- The NN learns to map from input to output by iteratively adjusting its neurons' parameters such that it minimizes the difference between its own output and the desired output using the training dataset.
- Backpropagation method: It allows the network errors to be projected back to every neuron's individual contribution and changes the parameters such that the error decreases the fastest (gradient descent) -> Iteratively using a random example from the training dataset to estimate such parameter change as opposed to evaluating over the entire training dataset (stochastic gradient descent; SGD).
- Over-fitting issue: The network performs extremely well on the training dataset but will generalize very poorly with the new unseen validation dataset -> Therefore, during training, the network performance is monitored using an unseen validation dataset and finally tested on an unseen dataset which was neither contained in training nor validation sets.
- Data augmentation: Generally, the training dataset should contain many different examples of the desired outputs, but networks used for microscopy applications are often trained with small examples (~thousands) to reach high prediction accuracy -> While data augmentation (generating features of different patterns from input images) can be a powerful way to supplement training datasets.
- In recent years, important technical developments have improved or sped up the learning stage.
- Transfer learning: The pre-trained networks allow much smaller training datasets to be used.
- Generative Adversarial Networks (GANs): One network learns to generate fake datasets, and another learns to discriminate fake from real.
- Self-learning or unsupervised learning: It allows the use of a very large non-curated dataset directly.
3. Neural networks and microscopy
3.1. Object detection and classification
- An important goal for microscopy image analysis is to recognize and assign identifies to relevant features on an image.
- Identifying mitotic cells in a tissue sample can be essential for cancer diagnosis.
- Classification NNs extensively used in the biomedical imaging field, especially for cancer detection, where it has shown expert-level recognition of subcellular features.
- A new approach (ex. Lu et al. [1]) in this area is to use unsupervised learning to identify subcellular protein localizations (it also removes the requirement for manual labeling of a training dataset).
- NNs have also shown their capacity to identify cellular states from transmitted-light data accurately.
- (1) Differentiating cells based on cell-cycle stage,
- (2) cells affected by phototoxicity,
- or (3) stem cell-derived endothelial cells.
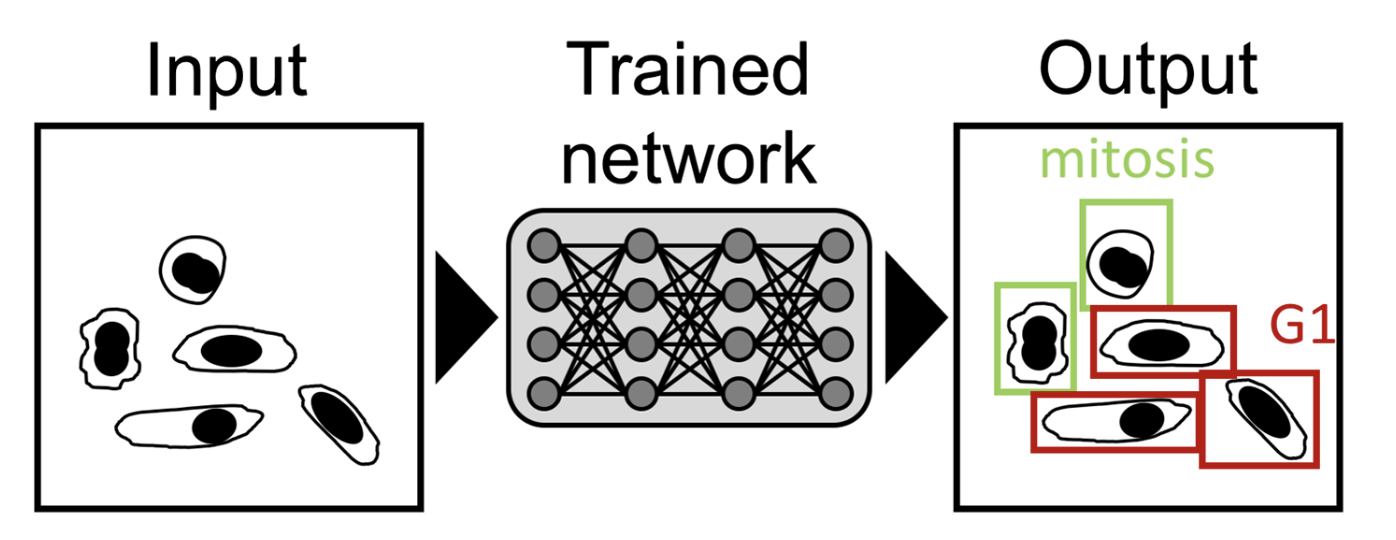
3.2. Image segmentation
- Segmentation is the identification of image regions that are part of specific cellular or subcellular structures and often is an essential step in image analysis.
- The NN identifies whether each pixel belongs to a category of structure (ex. background vs. signal).
- The CNNs have been successfully used to segment colon glands, breast tissues, and nuclei outperforming non-DL approaches.
- Segmentation is often used with subsequent classification and can even improve the accuracy of classification.

3.3. Artificial labeling
- While the task of artificial labeling is similar to segmentation, the main difference in this approach lies in the creation of the training dataset which does not require being hand-labeled.
- Instead, the training set contains paired images obtained from brightfield and fluorescence modalities of the same cells.
- The networks then learn to predict a fluorescent label from transmitted light or EM images, alleviating the need to acquire the corresponding fluorescence images.
- Christiansen et al. [2] also demonstrated their network's ability for transfer learning, allowing a pre-trained network to be applied between different microscopes and labels, highlighting the versatility of these networks' performance.

3.4. Image restoration: resolution and signal
- The amount and quality of features which can be extracted from a microscopic image are limited by fundamental constraints inherent to all optical set-ups: signal-to-noise ratio (SNR) and resolution.
- Overcoming these limitations constitutes a central goal in microscopy -> Super-Resolution Microscopy (SRM)
- However, phototoxicity, bleaching, and low temporal resolution still limit the capacity to achieve high-resolution long-term imaging in living specimens.
- For such networks, training datasets consist of, for instance, paired images acquired at low and high SNR, respectively and the network learns to predict a denoised (high SNR) image from a noisy input (low SNR).
- Weigert et al. [3] with their content-aware image restoration (CARE) methodology on the highly photosensitive organism Schmidtea Mediterranea which allowed a 60-fold decrease in illumination dose,
- Thus enabling longer and more detailed observation of this organism in vivo.
- Given the difficulties of creating large annotated training sets, different unsupervised learning methods for image restoration requiring no labeled training data have recently been explored.
- Here, a network learns image denoising on a dataset of noisy images alone.
3.5. Using neural networks in single-molecule localization microscopy
- Applying sophisticated network architectures, with combinations of widefield and SMLM data as inputs, the networks are able to directly map sparse SMLM data of either microtubule, mitochondria, or nuclear pores directly into their SRM output images.
- This demonstrates the strength of CNNs for pattern recognition in redundant data, like SMLM data where only a few frames may suffice to reconstruct an SRM image.
- Especially for high emitter density, this is advantageous over conventional SMLM reconstruction algorithms which can be time-consuming.
- Other studies have used a different approach to SMLM reconstruction in that networks are trained to detect the spatial positions of fluorophores from SMLM input images, similar to a typical SMLM algorithm.
- While achieving similar accuracy to state-of-the-art SMLM algorithms, the main achievement of DL for SMLM is the reconstruction speed with which super-resolved images can be produced.
* Reference: von Chamier, Lucas, Romain F. Laine, and Ricardo Henriques. "Artificial intelligence for microscopy: what you should know." Biochemical Society Transactions 47.4 (2019): 1029-1040.
- [1] Lu, A., Kraus, O.Z., Cooper, S. and Moses, A.M. (2018). Learning unsupervised feature representations for single cell microscopy images with paired cell inpainting. bioRxiv, 395954 https://doi.org/10.1101/395954
Learning unsupervised feature representations for single cell microscopy images with paired cell inpainting
Cellular microscopy images contain rich insights about biology. To extract this information, researchers use features, or measurements of the patterns of interest in the images. Here, we introduce a convolutional neural network (CNN) to automatically desig
www.biorxiv.org
- [2] Christiansen, E.M., Yang, S.J., Ando, D.M., Javaherian, A., Skibinski, G., Lipnick, S., et al. (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173, 792–803.e19 https://doi.org/10.1016/j.cell.2018.03.040
Redirecting
linkinghub.elsevier.com
- [3] Weigert, M., Schmidt, U., Boothe, T., Müller, A., Dibrov, A., Jain, A., et al. (2018) Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 https://doi.org/10.1038/s41592-018-0216-7




댓글