# Three-line Summary #
- Single-molecule localization microscopy (SMLM) has had remarkable success in imaging cellular structures with nanometer resolution, but standard analysis algorithms require sparse emitters, which limits imaging speed and labeling density.
- We developed DECODE (deep context dependent; using deep learning), a computational tool that can localize single emitters at high density in three dimensions with the highest accuracy for a large range of imaging modalities and conditions.
- DECODE allowed us to acquire fast dynamic live-cell SMLM data with reduced light exposure and to image microtubules at ultra-high labeling density.
# Detail Review #
1. Introduction
- Single-molecule localization microscopy (SMLM; ex. PALM, STORM) has become an invaluable super-resolution method for biology, but it is based on acquiring a large number of camera frames.
- Only a tiny fraction of the emitters are stochastically activated into a bright 'on' state so that their images do not overlap.
- This allows precise localization of the emitter coordinates by fitting a model of the point spread function (PSF), and a super-resolution image is then reconstructed from these coordinates.
- This principle of SMLM is at the same time one of its main limitations: the need for sparse activation leads to long acquisition times.
- For SMLM, deep learning holds promise to extract emitter coordinates and additional parameters under conditions and densities too complex for traditional fitters.
- While ground-truth data to train the neural network are typically not available, synthetic training data can be generated by numerically simulating the imaging process.
- Here we present the DECODE (deep context-dependent) method for deep learning-based single-molecule localization that achieves high accuracy across a wide range of emitter densities and brightness levels.
- In the public SMLM challenge, DECODE outperformed all existing methods on 12 out of 12 datasets.
- DECODE enables fast live-cell SMLM with reduced light exposure and visualization of dynamic processes.
2. Method
- Architecture: DECODE introduces a new output representation and architecture for detecting and localizing emitters that predicts multiple channels with the same dimensions as the input image.
- Predictions: (1) Pixel probability of detecting an emitter, (2) emitter's subpixel spatial coordinates, (3) brightness, (4) uncertainty related to those predictions, (5) optional background map
- To overcome the limitations of previous SMLM methods: (1) DECODE predictions scale only with the number of image pixels, which improves prediction speeds and eliminates a voxel size-dependent limit on precision. (2) DECODE has four additional output channels that estimate the uncertainty of the localization along each coordinate (x, y, z) and the brightness. (3) DECODE network integrates information across multiple frames with a two-stage design: 'Frame analysis module (U-Net)' for single imaging frames & 'Temporal context module (U-Net)' for integrating feature representations of the frames (previous frame, current frame, next frame)
- Dataset: we trained the DECODE network by generating a large amount of simulated data
- Ground truth data for supervised learning are not easily available for SMLM.
- It is possible to simulate realistic images of activated emitters as the physics of imaging single molecules is well understood.
- Training loss: we trained the DECODE network to predict probability of detection, along with the subpixel localization and localization uncertainty of each detected emitter.
- (1) Count loss: compares the true and detected number of emitters in the images.
- (2) Localization loss: trains the network to correctly localize the detected emitters and estimate the localization uncertainty and emitter brightness
- (3) Optional background loss: computes the mean squared error between the true and predicted background images.

3. Result
- DECODE achieves high accuracy for a wide range of simulated data.
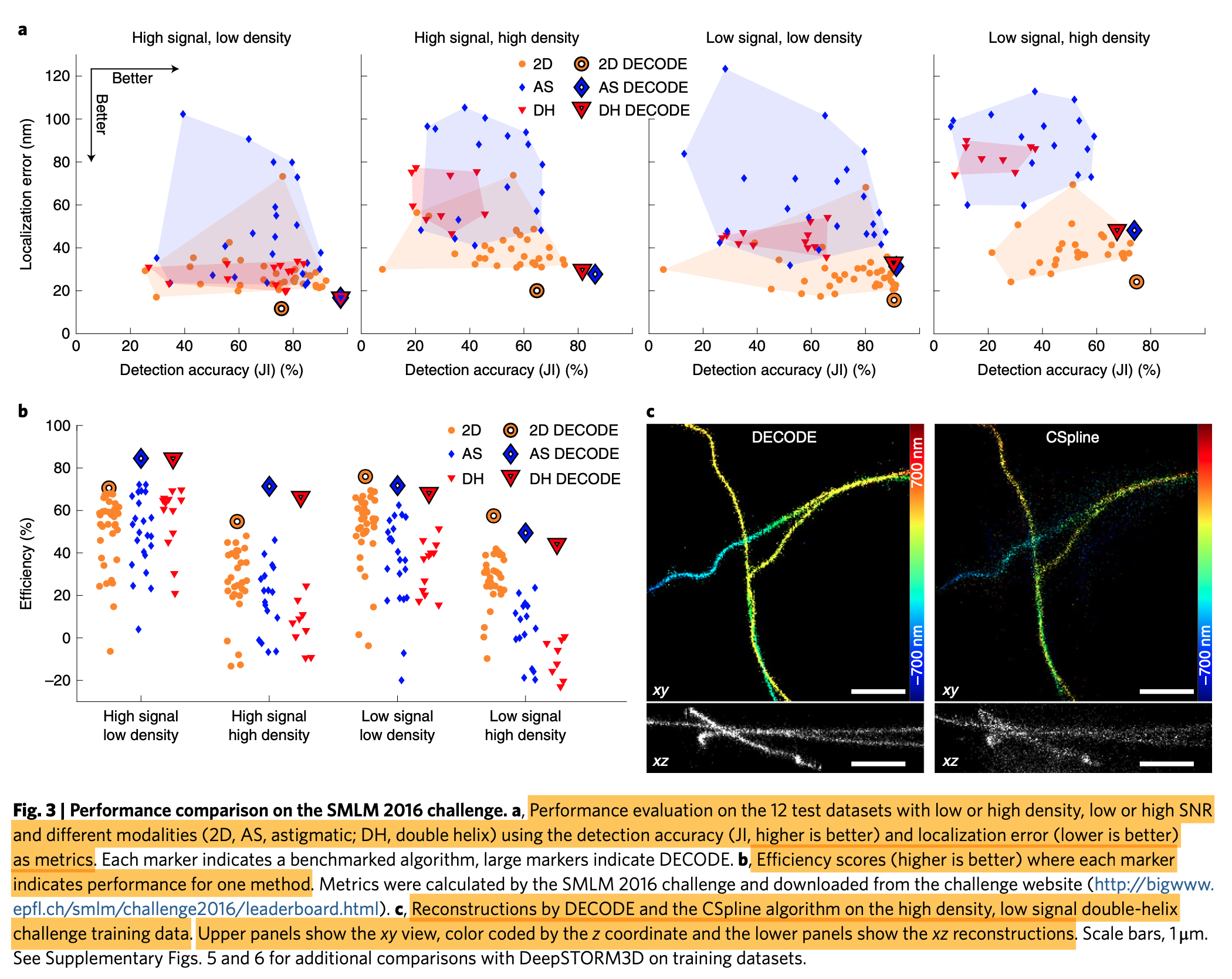
- Performance metrics: (1) the detection accuracy quantifies the fraction of emitters that are detected (Jaccard Index; JI = TP/(TP + FN + FP)), (2) localization error, that is how close the measured coordinates are to the true coordinates, measured here as the root mean squared error (r.m.s.e.) averaged over the dimensions.
- There is a natural trade-off between JI and localization error: discarding all but the brightest and best-separated emitters will result in a good (low) localization error but a bad (low) JI.
- Simulated data result: (1) DECODE approaches the CRLB for low densities. (2) DECODE's uncertainty estimates are well-calibrated. (3) Temporal context improves localization error and detection. (4) DECODE architecture outperforms a voxel-based network architecture and a multi-emitter fitter.

- DECODE outperforms all fitters on a public SMLM benchmark (SMLM 2016 challenge).
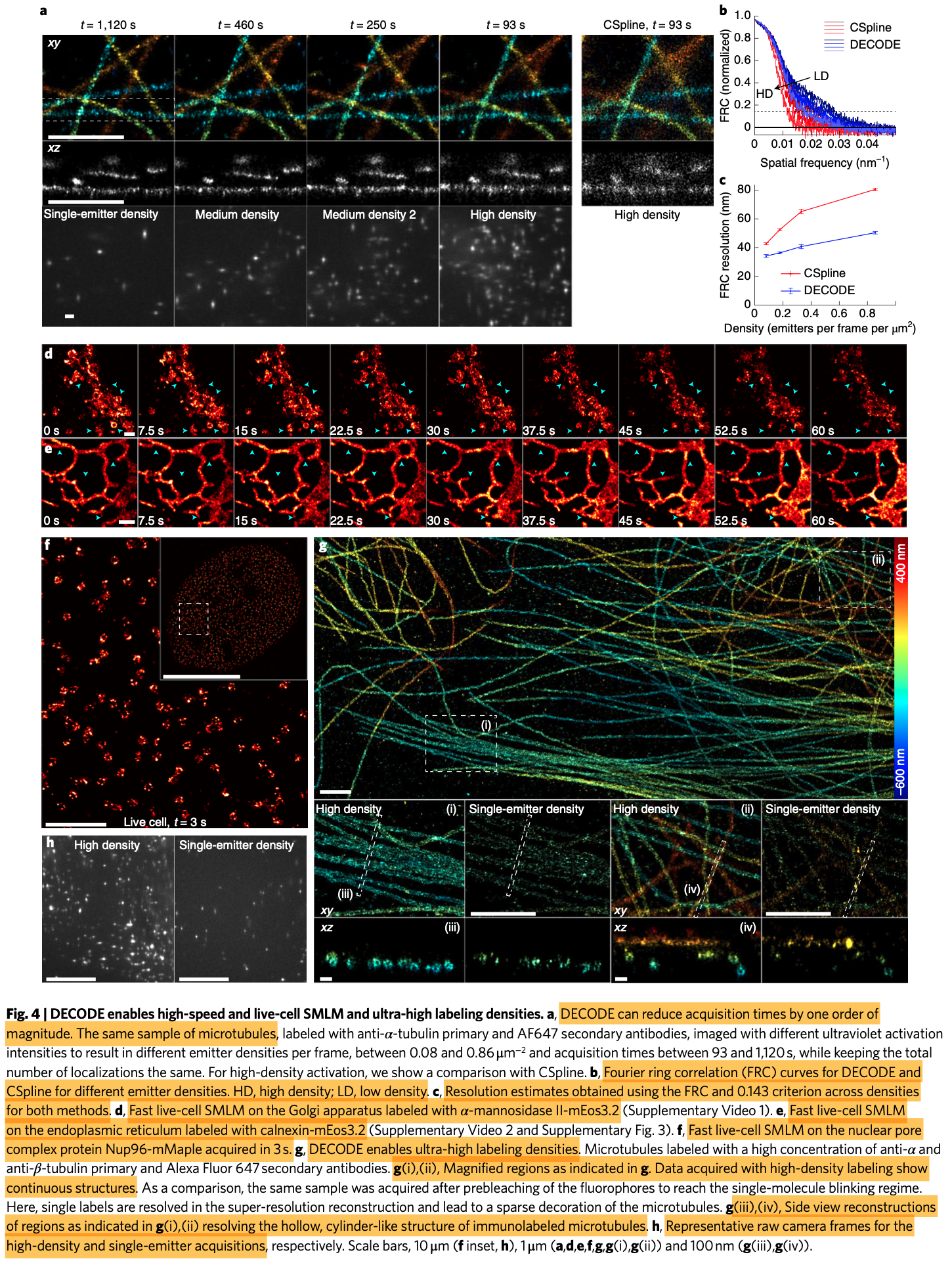
- DECODE reduces imaging times by one order of magnitude, enabling fast live-cell SMLM with reduced light exposure.
* Reference: Speiser, Artur, et al. "Deep learning enables fast and dense single-molecule localization with high accuracy." Nature methods 18.9 (2021): 1082-1090.




댓글