# Three-line Summary #
- Microscopy images typically vary in signal-to-noise ratios and include a wealth of information that requires multiple parameters and time-consuming iterative algorithms for processing, but deep learning technologies develop quickly, and they have been applied in bioimage processing more and more frequently.
- This review article introduces the applications of deep learning algorithms in microscopy image analysis, which include image classification, region segmentation, object tracking, and super-resolution reconstruction.
- Furthermore, the latest development of augmented intelligent microscopy that is based on deep learning technology may lead to a revolution in biomedical research.
# Detail Review #
1. Introduction
- With the developments of optics and computer science, advanced technologies of microscopy have opened up new eyesight for biomedical researchers.
- Fluorescence microscopy, such as confocal and total internal reflection fluorescence microscopy (TIRFM) have been widely used in biomedical research to observe subcellular structures with specific labeling.
- Super-resolution microscopy is a new trend of microscope development that breaks the diffraction limit and records biological processes at the nanometer scale.
- But due to imaging constraints and diffraction limitation, conventional fluorescence microscopy images always have relatively low resolution and poor signal-to-noise, for which traditional image analysis methods could not have a robust performance.
- Nowadays, deep learning has developed and evolved in all aspects of scientific fields, especially in the field of image processing.
- Different from traditional image processing methods, one of the key advantages of deep learning is that its layers of features are not designed by humans.
- Deep learning is able to learn from end to end itself and does not require complex manual computation but result annotation instead.
- This review is organized in order of applications of deep learning in microscopy image analysis, and we categorize them into 4 groups according to the main research targets.
- Applications of deep learning in microscopy image analysis: Classification, Segmentation, Tracking, Reconstruction
- As for SMLM, CNN-based single-molecule localization models can be helpful to speed up the data-processing procedure with robustness.
- Intelligent augmented microscopy: combines microscopy with deep learning, enables super-resolution imaging and high-content, efficient, real-time analysis.


2. Classification
2.1. Deep learning-based classifiers
- As deep learning develops, neural network classifiers, especially those based on convolution neural networks (CNN), are growing in popularity.
- Most of the CNN models contain two parts, the feature extraction module consisting of convolution and pooling layers and the classification module consisting of fully-connected layers. (AlexNet, VggNet, GoogLeNet, ResNet).
2.2. Applications of deep learning-based classifiers
- Cellular and subcellular classification: deep learning-based classifiers were able to identify different types of cells and cells at different stages of differentiation with high accuracy.
- Disease diagnosis: Researchers have proposed several classifiers to identify white blood cells or other human diseases (lung cancer subtypes, diagnosis of hepatic granuloma, breast cancer, colon cancer).

3. Segmentation
3.1. Deep learning-based segmentation methods
- Image segmentation is the process of dividing an image into several regions with certain properties that people are interested in that can be divided into semantic-level segmentation and instance-level segmentation.
- Semantic segmentation classifies each pixel in an image into the foreground and background (FCN, U-Net).
- Instance-level segmentation is based on target detection, which identifies different objects in the image and classifies them (R-CNN, Fast R-CNN, Faster R-CNN, Mask R-CNN).
3.2. Applications of deep learning-based segmentation methods
- Diagnose for diseases: Methods for highly accurate cell detection and segmentation are greatly needed in drug discovery and cancer research.
- Intracellular compartment segmentation: Besides segmentation on a single-cell scale, deep learning also performs well on the subcellular and tissue scale.
- Tissue image analysis: Segmentation methods based on deep learning also play an important role in tissue image analysis.
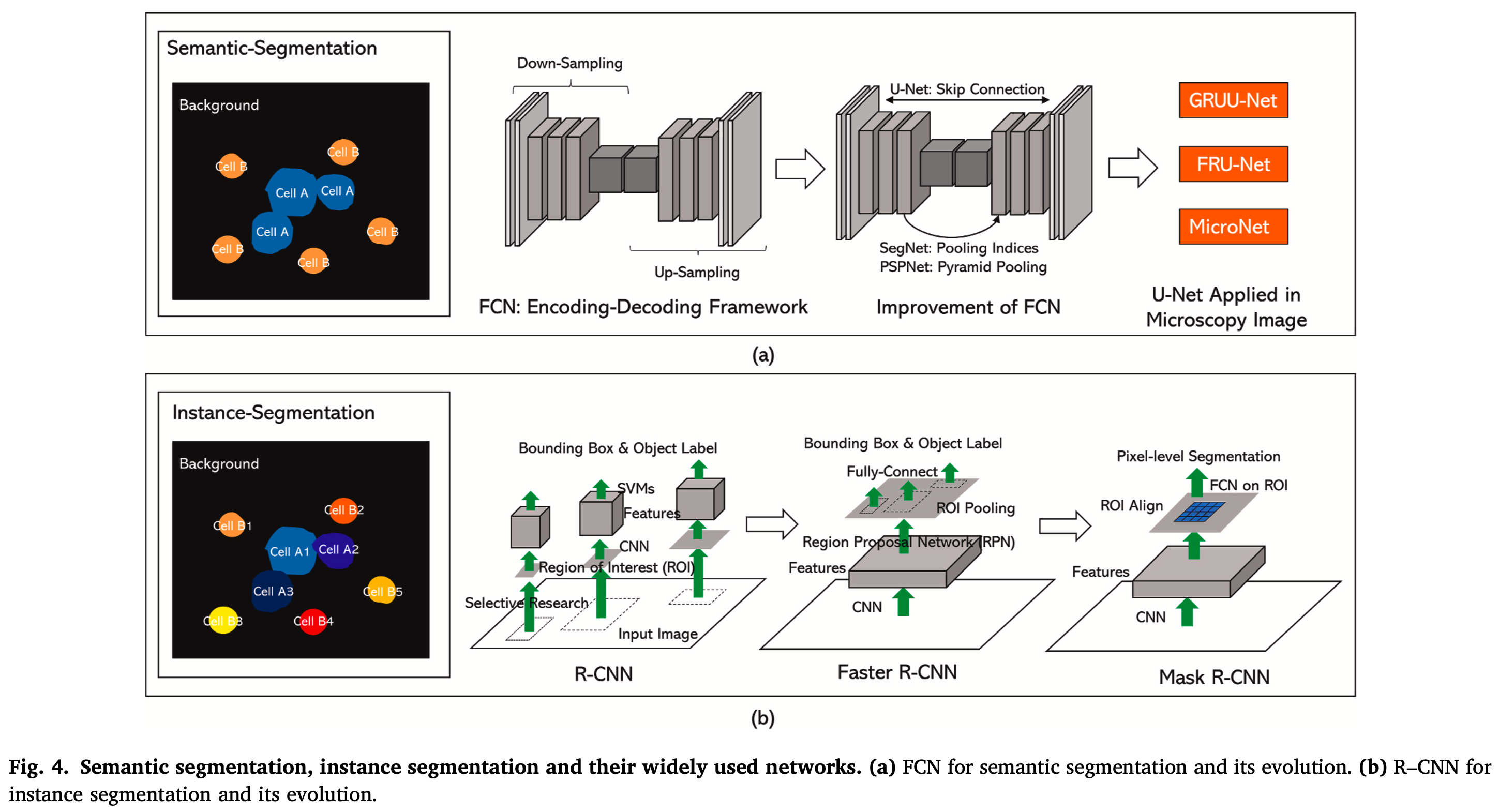
4. Tracking
4.1. Deep learning-based object tracking
- Object tracking is the task of following objects through a series of time-lapse images that tasks can be divided into two steps, instance-level localization and data association.
- The most commonly used networks for localization used by Mask R-CNN (used to segment and track nucleus and microtubule in a cell) and data association used by RNN + LSTM (preserve previous information and enable the model to memorize).
4.2. Applications of deep learning-based object tracking in microscopy image
- Cell tracking:
- To determine the drug treatment effects on cancer cells,
- To perform rapid antibiotic susceptibility testing
- To analyze tumor cell metastasis, wound healing, and neural crest migration.
- Intracellular particle tracking: A deep learning-based software for automated kymograph analysis called KymoButler was developed to visualize the dynamics of fluorescent particles, molecules, vesicles, and organelles along a predictable trajectory.

5. Reconstruction
5.1. Deep learning-based image reconstruction
- Conventional super-resolution microscopy
- (1) The modification of point spread functions (PSFs): structured illumination microscopy (SIM) & stimulated emission depletion microscopy (STED).
- (2) Single-molecule localization and complicated imaging analysis algorithm: photoactivated localization microscopy (PALM) & stochastic optical reconstruction microscopy (STORM) & super-resolution radial fluctuations (SRRF).
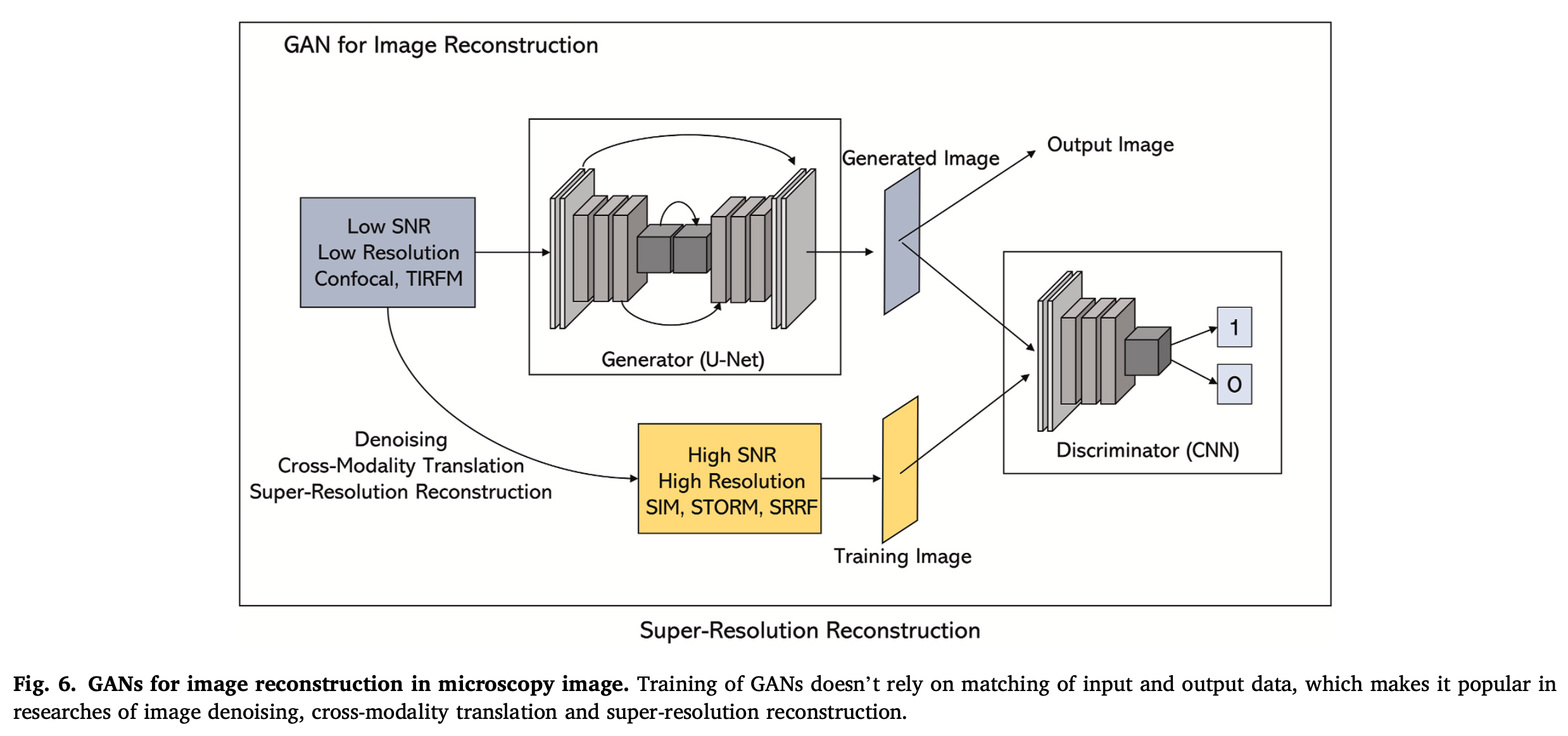
- Apart from traditional convolution network, GANs is a promising model for microscopy image reconstruction that consists of two parts,
- A generator (usually an FCN or U-Net, which is used to generate an image)
- A discriminator (usually a CNN, which is used to discriminate whether the generated image is real or fake).
5.2. Applications of deep learning-based reconstruction in microscopy image
- Optimization of the imaging parameters, image denoising & image restoration:
- The fluorescent-labeled biological samples usually vary in quality, thus one would use different laser power, scanning speed, and exposure times to meet the requirements
- Deep learning networks have been used to automatically set the parameters for imaging, which enable the imaging system to be adaptive.
- Acceleration of imaging speed:
- single-molecule localization microscopy (SMLM) sacrifices temporal resolution for spatial resolution and has limitations on the visualization of live cells.
- But CNN-based single-molecule localization models can be helpful to speed up the data-processing procedure with robustness (ex. Deep-STORM, ANNA-PALM).
- Cross-modality super-resolution reconstruction:
- The GANs have been used to enable super-resolution imaging across different microscope systems (image-to-image translation), that is, converting diffraction-limited input images to super-resolved ones.
* Reference: Liu, Zhichao, et al. "A survey on applications of deep learning in microscopy image analysis." Computers in Biology and Medicine 134 (2021): 104523.




댓글